Pfam MySQL database¶
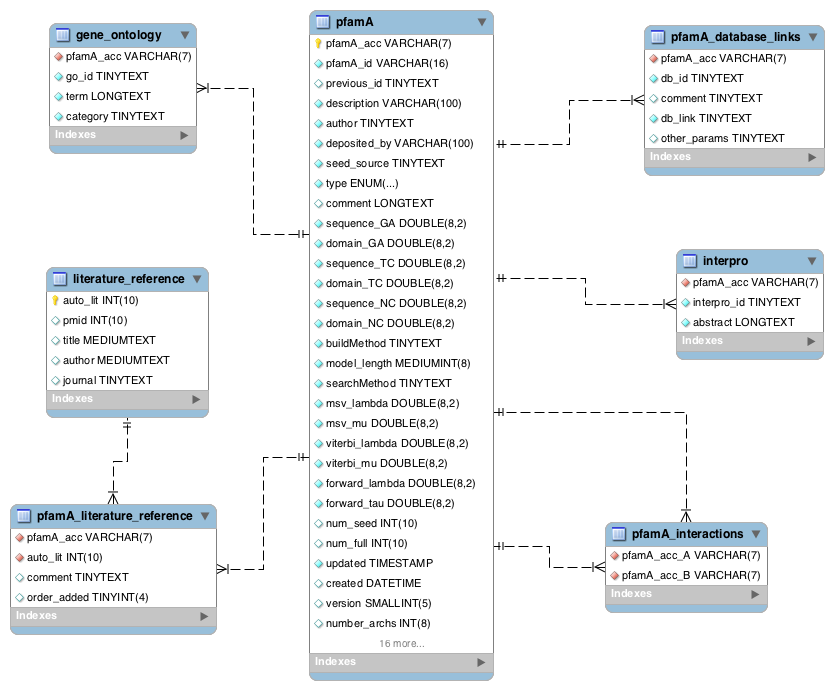
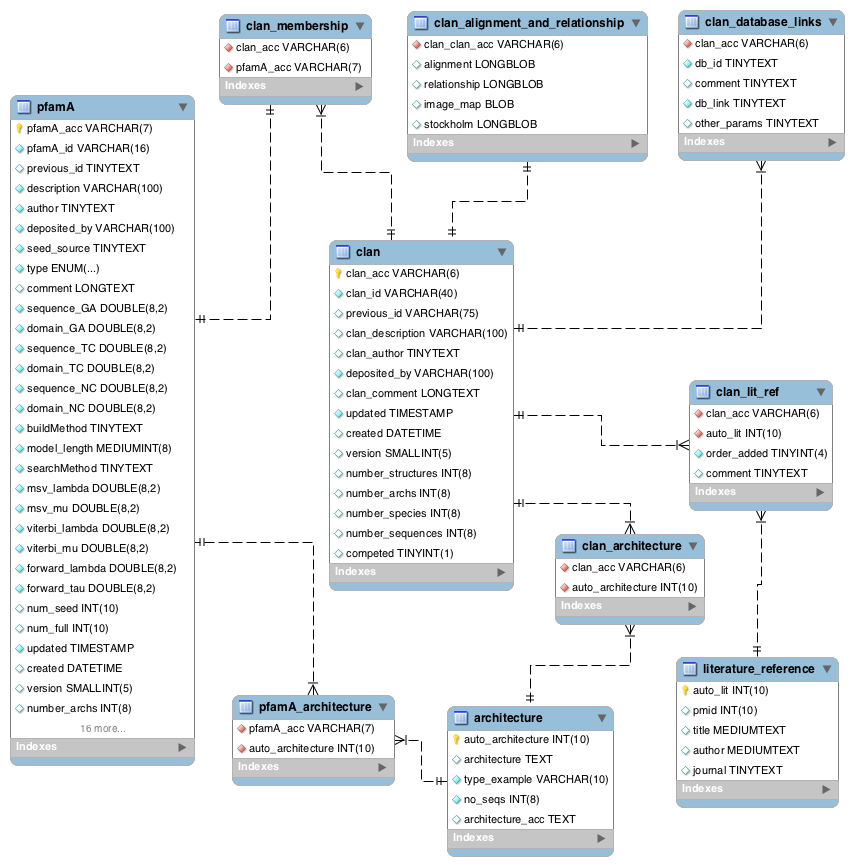
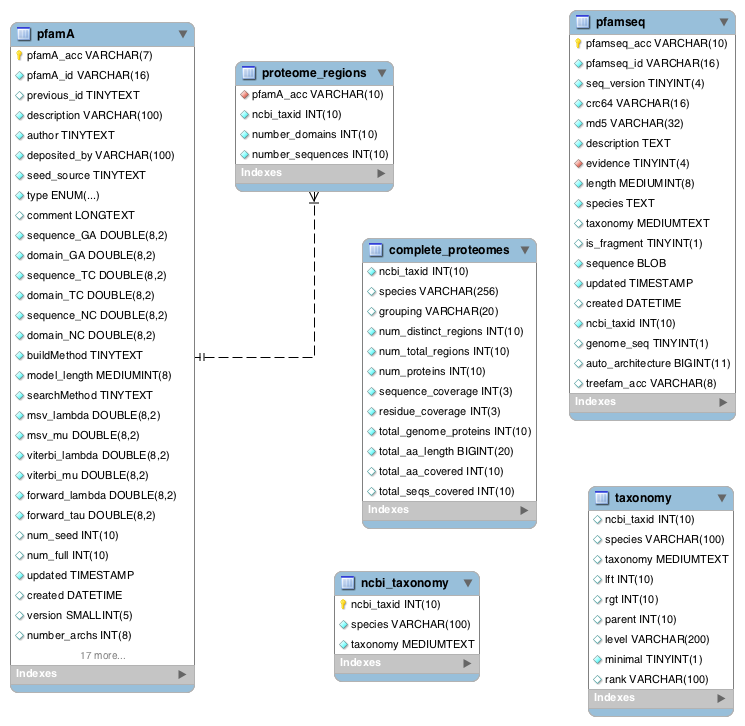
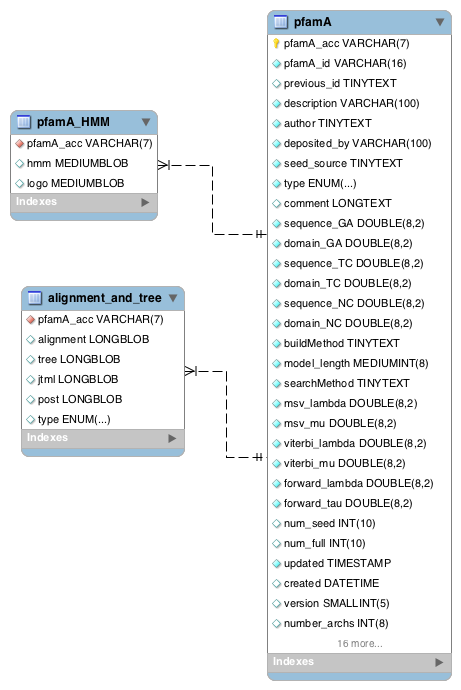
The Pfam MySQL database contains all of the data accessible via the website. The database currently consists of 63 tables. Below is some basic documentation on the schema layout and how smaller numbers of tables can be put together to enable access to a subset of the data. At the time of writing, release 36.0, the fields within the tables and the results of queries are correct. The data within the tables will change with each release. Although we do not anticipate any major changes to the database, we reserve the right to make changes with or without warning; we will endeavour to update this document if such changes are made. Please note that we do not provide a public version of the Pfam database after release 36.0.
A red diamond in the images below indicates a foreign key. In some images there are tables which appear not to be linked to any other table in the image. This is due to a foreign key being populated late in production of the database. The ‘floating’ table can still be joined and example queries of how to do so are given under each image.
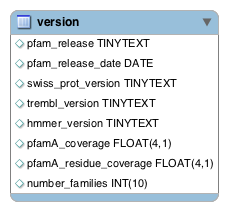
Version table¶
The version table contains information that relates to a particular Pfam release. It contains the version number of the Pfam database, the version numbers of the Swiss-Prot and TrEMBL databases that were used to build Pfam, and some statistics about the number of families and coverage. This table is stand-alone and does not link to any of the other tables.
Example query: Give me all of the version information for the Pfam database
SELECT *
FROM version
Example output:
pfam_release: 36.0
pfam_release_date: 2023-07-18
swiss_prot_version: 2022_05
trembl_version: 2022_05
hmmer_version: 3.3
pfamA_coverage: 76.2
pfamA_residue_coverage: 48.6
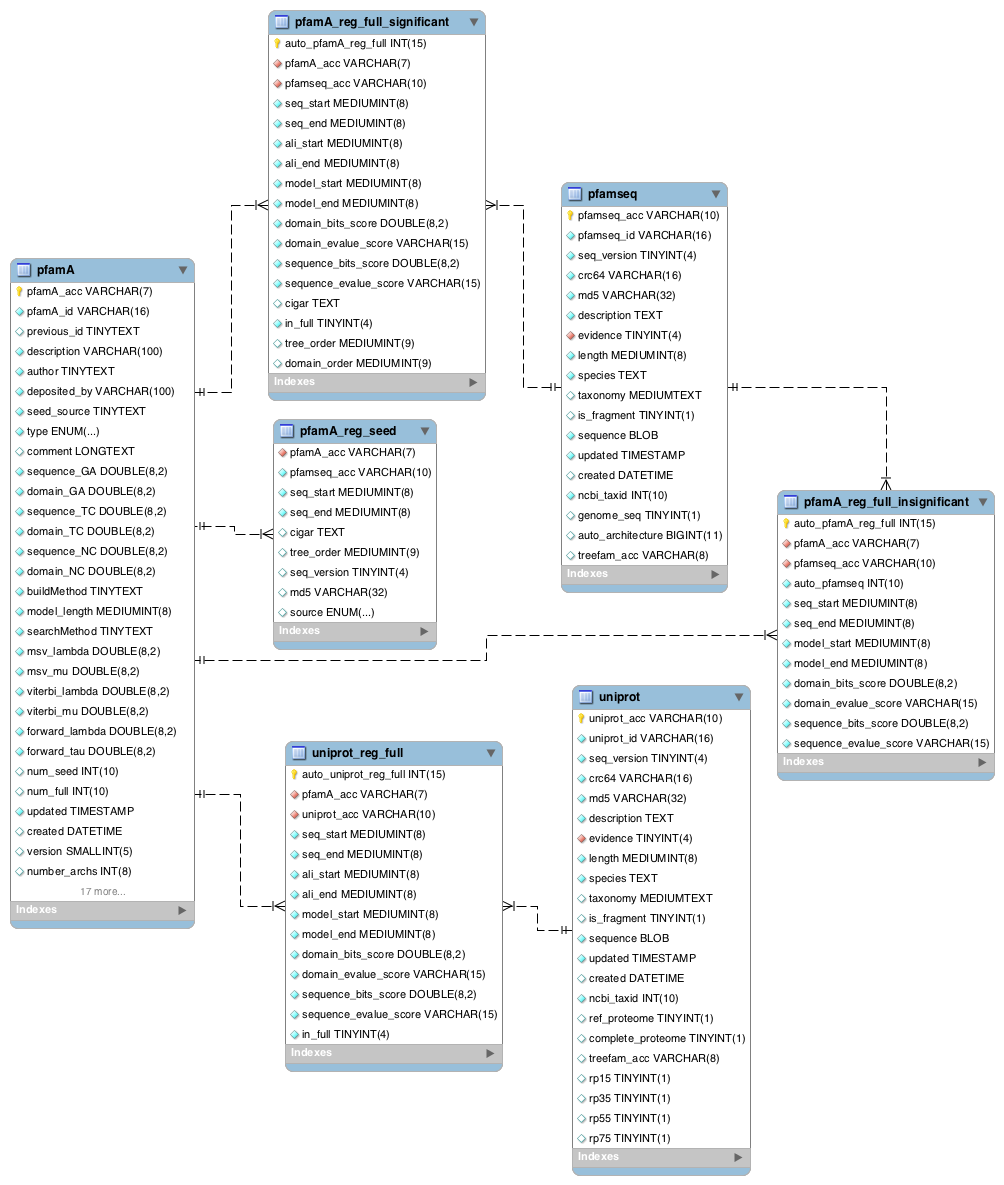
Domain information¶
Two of the central tables in the Pfam database are pfamseq, which contains UniProtKB reference proteomes and pfamA, which contains information about the Pfam-A families. Most of the other tables in the database link to one or both of these tables, either directly or indirectly. Note that prior to Pfam 29.0, the pfamseq table contained the whole of UniProtKB. From Pfam 29.0, this table contains only the reference proteome portion of UniProtKB. The full alignments in Pfam are based on the sequences in the pfamseq table.
The table pfamA_reg_seed contains the Pfam regions that are present in a seed alignment. All sequences in pfamA_reg_seed are in the pfamseq table or the uniprot table (the uniprot table contains all the sequences in UniProtKB). The pfamA_reg_full_significant table contains all of the sequence regions from the pfamseq table that match the HMM and score above the curated threshold, i.e. are significant matches, for each family. There is also a table named pfamA_reg_full_insignificant which contains, as the name suggests, all the insignificant matches for each family. Insignificant matches are those which match the HMM with an E-value less than 1000, but score below the curated bit score threshold for each family.
In addition to providing matches to the sequences in the pfamseq table, we also provide the significant matches for the sequences in the uniprot table. These can be found in the table uniprot_reg_full.
The tables pfamA_reg_full_significant and uniprot_reg_full contain a column called ‘in_full’. The matches that are present in the full alignment for a Pfam family have this column set to 1, while those that are not present in the full alignment have the ‘in_full’ column set to 0. A significant match will only be excluded from the full alignment (in_full = 0) if it matches a family that belongs to a clan, and the match overlaps with another more significant (lower E-value) match to a family within the clan.
For each sequence match we store two sets of coordinates, the envelope coordinates and the alignment coordinates. The envelope co-ordinates delineate the region on the sequence where the match has been probabilistically determined to lie, whereas the alignment coordinates delineate where HMMER is confident that the alignment of the sequence to the profile HMM is correct. Our full alignments contain the envelope coordinates. In the database, envelope start and end positions are stored in the seq_start and seq_end fields columns, and the alignment coordinates are stored in the ali_start and ali_end fields.
The Pfam database has historically been built on the UniProtKB database. However, as of release 22.0 we also provide Pfam domain data for the NCBI sequence database (GenPept) and a set of metagenomics sequences. As of release 28.0, we no longer store Pfam information at the sequence level for the NCBI and metagenomics data sets in the MySQL database, but we still provide the family alignments for them in the alignment_and_tree table.
Example query: Give me all of the domains for sequence ‘VAV_HUMAN’
SELECT pfamA.pfamA_acc, pfamA_id, seq_start, seq_end
FROM pfamseq, pfamA, pfamA_reg_full_significant
WHERE pfamseq_id = 'VAV_HUMAN'
AND in_full = 1
AND pfamseq.pfamseq_acc = pfamA_reg_full_significant.pfamseq_acc
AND pfamA_reg_full_significant.pfamA_acc = pfamA.pfamA_acc
ORDER BY seq_start
Example output:
pfamA_acc pfamA_id seq_start seq_end
PF00307 CH 1 121
PF00621 RhoGEF 198 371
PF00169 PH 403 504
PF00130 C1_1 516 568
PF00018 SH3_1 615 652
PF00017 SH2 671 745
PF00018 SH3_1 788 834
To report all of the overlapping domains within any clans, leave out the ‘in_full =1’ clause. More information on clans can be found later in this document.
Example query: Give me all the sequences in the full alignment for the family ‘B12D’
SELECT pfamseq_id, pfamseq.pfamseq_acc, seq_start, seq_end, pfamA_id
FROM pfamA, pfamseq, pfamA_reg_full_significant
WHERE pfamA_id = 'B12D'
AND in_full = 1
AND pfamA.pfamA_acc = pfamA_reg_full_significant.pfamA_acc
AND pfamA_reg_full_significant.pfamseq_acc = pfamseq.pfamseq_acc
Example output:
pfamseq_id pfamseq_acc seq_start seq_end pfamA_id
A0A4W6ED51_LATCA A0A4W6ED51 11 80 B12D
A0A2C6L090_9APIC A0A2C6L090 43 96 B12D
A0A1U8P0E7_GOSHI A0A1U8P0E7 5 66 B12D
A0A6P6JK09_CARAU A0A6P6JK09 11 79 B12D
A0A436ZUI8_9PEZI A0A436ZUI8 55 96 B12D
A0A0D9WNU3_9ORYZ A0A0D9WNU3 8 76 B12D
A0A5N5FAS6_9ROSA A0A5N5FAS6 7 38 B12D
A0A1V4KBT8_PATFA A0A1V4KBT8 11 79 B12D
B6DDV9_ANODA B6DDV9 11 78 B12D
A0A0C9M2H1_9FUNG A0A0C9M2H1 19 86 B12D
...
Example query: Give me all the sequences in the seed alignment for the family ‘B12D’
SELECT pfamseq_id, pfamseq.pfamseq_acc, seq_start, seq_end, pfamA_id
FROM pfamA, pfamseq, pfamA_reg_seed
WHERE pfamA_id = 'B12D'
AND pfamA.pfamA_acc = pfamA_reg_seed.pfamA_acc
AND pfamA_reg_seed.pfamseq_acc = pfamseq.pfamseq_acc
Example output:
pfamseq_id pfamseq_acc seq_start seq_end pfamA_id
M3ZJB0_XIPMA M3ZJB0 36 105 B12D
M1C9M8_SOLTU M1C9M8 7 75 B12D
E2B831_HARSA E2B831 11 78 B12D
A0A4S8III1_MUSBA A0A4S8III1 5 73 B12D
I1H296_BRADI I1H296 6 74 B12D
K7DYW7_MONDO K7DYW7 9 77 B12D
I1H293_BRADI I1H293 14 82 B12D
F4WMZ4_ACREC F4WMZ4 11 78 B12D
J3MJY8_ORYBR J3MJY8 17 85 B12D
A0A804JLQ3_MUSAM A0A804JLQ3 5 73 B12D
...
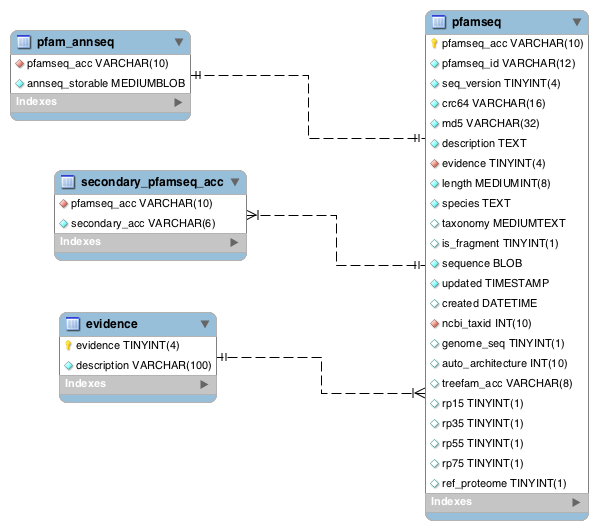
Pfamseq - other tables¶
This section contains a few tables that link to the pfamseq table, but don’t fit nicely into any of the sections described above.
The evidence table contains the UniProtKB evidence code key that is used in the evidence field in the pfamseq and uniprot tables.
UniProtKB sequences have secondary accessions if they have been merged or split. Secondary accession numbers are stored in the table called secondary_pfamseq_acc.
Example query: Give me the secondary accession(s) for the sequence ‘P15455’
SELECT secondary_acc
FROM pfamseq, secondary_pfamseq_acc
WHERE pfamseq.pfamseq_acc = secondary_pfamseq_acc.pfamseq_acc
AND pfamseq.pfamseq_acc= 'P15455'
Example output:
secondary_acc
Q3E711
Q56Z11
Q9FFH7
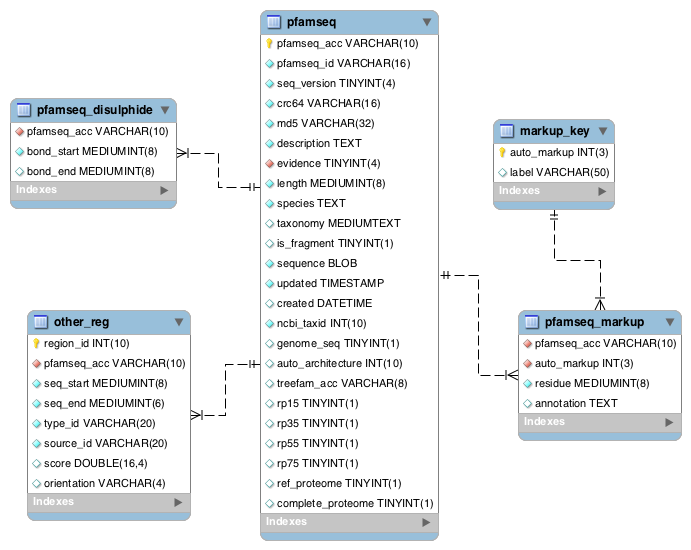
Other regions¶
These tables contain sequence specific information about the reference proteome sequences. The other_regions table contains coiled coil, low complexity, signal peptide, transmembrane and disordered regions data. The pfamseq_markup table contains active site information which is taken from the UniProtKB feature table. Additional active site residues are predicted by Pfam based on conserved residues in a Pfam alignment. The pfamseq_disulphide table contains disulphide bond information from the UniProtKB feature table.
Example query: Give me all of the transmembrane, signal-peptide, coiled-coils, low-complexity and disorder information for the sequence ‘VAV_HUMAN’
SELECT type_id, source_id, seq_start, seq_end
FROM other_reg, pfamseq
WHERE pfamseq.pfamseq_id = 'VAV_HUMAN'
AND other_reg.pfamseq_acc = pfamseq.pfamseq_acc
Example output:
type_id source_id seq_start seq_end
disorder IUPred 128 129
disorder IUPred 140 141
disorder IUPred 160 161
disorder IUPred 173 177
disorder IUPred 179 180
disorder IUPred 568 588
disorder IUPred 635 636
low_complexity segmasker 42 51
low_complexity segmasker 356 367
Example query: Give me all of the active site information for sequence ‘F7PG13’
SELECT pfamseq.pfamseq_acc, pfamseq_id, residue, label
FROM pfamseq, pfamseq_markup, markup_key
WHERE pfamseq.pfamseq_acc = pfamseq_markup.pfamseq_acc
AND pfamseq_markup.auto_markup = markup_key.auto_markup
AND pfamseq.pfamseq_acc = 'F7PG13'
Example output:
pfamseq_acc pfamseq_id residue label
F7PG13 F7PG13_9EURY 92 Pfam predicted active site
F7PG13 F7PG13_9EURY 248 Pfam predicted active site
F7PG13 F7PG13_9EURY 92 UniProt predicted active site
Example query: Give me all the residues involved in disulphide bonds in the sequence ‘Q43495’
SELECT pfamseq.pfamseq_acc, pfamseq_id, bond_start, bond_end
FROM pfamseq, pfamseq_disulphide
WHERE pfamseq_disulphide.pfamseq_acc = pfamseq.pfamseq_acc
AND pfamseq.pfamseq_acc = 'Q43495'
Example output:
pfamseq_acc pfamseq_id bond_start bond_end
Q43495 108_SOLLC 41 77
Q43495 108_SOLLC 79 99
Q43495 108_SOLLC 51 66
Q43495 108_SOLLC 67 92
Annotation information for a family¶
In addition to the Pfam annotation, we also store InterPro annotation and their associated GO terms for each family. Links to other databases, e.g. SCOP) are also stored where appropriate. The pfamA table contains the GA, TC and NC cut-offs for each family, and additional information surrounding the Pfam-A family, including the number of sequences in the seed and full alignment. The pfamA_interactions table contains, where data are available, pairs of interacting Pfam domains. The data in this table are taken from the iPfam resource (no longer maintained), which describes physical interactions between Pfam domains that have a representative structure in the PDB.
Example query: Give me the Pfam annotation for the family ‘CBS’
SELECT comment
FROM pfamA WHERE pfamA_id = 'CBS'
Example output:
comment: CBS domains are small intracellular modules that pair together
to form a stable globular domain [2]. This family represents a single CBS
domain. Pairs of these domains have been termed a Bateman domain [6]. CBS
domains have been shown to bind ligands with an adenosyl group such as
AMP, ATP and S-AdoMet [5]. CBS domains are found attached to a wide
range of other protein domains suggesting that CBS domains may play a
regulatory role making proteins sensitive to adenosyl carrying ligands.
The region containing the CBS domains in Cystathionine-beta synthase is
involved in regulation by S-AdoMet [4]. CBS domain pairs from AMPK bind
AMP or ATP [5]. The CBS domains from IMPDH and the chloride channel CLC2
bind ATP [5].
Example query: Give me all of the literature references for the family ‘CBS’
SELECT pfamA_literature_reference.comment, order_added, pmid, title, literature_reference.author, journal
FROM pfamA, pfamA_literature_reference, literature_reference
WHERE pfamA_id = 'CBS'
AND pfamA.pfamA_acc = pfamA_literature_reference.pfamA_acc
AND pfamA_literature_reference.auto_lit = literature_reference.auto_lit
Example output:
comment: Discovery and naming of the CBS domain.
order_added: 1
pmid: 9020585
title: The structure of a domain common to archaebacteria and the homocystinuria disease protein.
author: Bateman A;
journal: Trends Biochem Sci 1997;22:12-13.
...
Example query: Give me all of the database references for the family ‘A2M’
SELECT db_id, pfamA_database_links.comment, db_link, other_params
FROM pfamA, pfamA_database_links
WHERE pfamA_id = 'A2M'
AND pfamA.pfamA_acc = pfamA_database_links.pfamA_acc
Example output:
db_id comment db_link other_params
PROSITE PDOC00440
SCOP 1c3d fa
HOMSTRAD A2M_A
HOMSTRAD A2M_B
Note: The other_params column contains ‘fa;’ where the Pfam family corresponds to a SCOP family, and ‘sf;’ where the Pfam family corresponds to a SCOP superfamily.
Clan data¶
A Pfam clan is a set of related Pfam-A families. The information we use to determine which families belong to the same clan includes related structure, related function, matching of the same sequence to HMMs from different families, and profile-profile comparisons. Note that not all Pfam-A families belong to a clan and that a Pfam-A family cannot belong to more than one clan.
Example query: Give me the id and accession of the clan to which Pfam family ‘EGF’ belongs
SELECT clan_id, clan.clan_acc
FROM clan, clan_membership, pfamA
WHERE clan.clan_acc = clan_membership.clan_acc
AND clan_membership.pfamA_acc = pfamA.pfamA_acc
AND pfamA.pfamA_id = 'EGF'
Example output:
clan_id clan_acc
EGF CL0001
Example query: Give me all of the Pfam-A families that belong to clan ‘CL0001’
SELECT pfamA.pfamA_acc, pfamA_id
FROM clan, clan_membership, pfamA
WHERE clan.clan_acc = clan_membership.clan_acc
AND clan_membership.pfamA_acc = pfamA.pfamA_acc
AND clan.clan_acc = 'CL0001'
Example output:
PF01414 DSL
PF04863 EGF_alliinase
PF00053 Laminin_EGF
PF07645 EGF_CA
PF00008 EGF
PF07974 EGF_2
PF09064 Tme5_EGF_like
PF09289 FOLN
PF12661 hEGF
PF12662 cEGF
PF12946 EGF_MSP1_1
PF12947 EGF_3
PF14670 FXa_inhibition
PF06247 Plasmod_Pvs28
PF09443 CFC
PF00084 Sushi
PF09014 Sushi_2
PF00594 Gla
PF18193 Fibrillin_U_N
PF18372 I-EGF_1
PF18720 EGF_Tenascin
PF07699 Ephrin_rec_like
PF20626 Sp38_C
PF21195 C8A_B_C6_EGF-like
PF21284 C7_FIM2_N
PF21286 CFAI_FIMAC_N
PF21364 FBN_EGF_st1
PF21700 DL-JAG_EGF-like
PF21795 JAG1-like_EGF2
Example query: Give me the clan description and comment for clan ‘CL0001’
SELECT clan.clan_acc, clan_id, clan_description, clan_comment
FROM clan
WHERE clan_acc = 'CL0001'
Example output:
clan_acc: CL0001
clan_id: EGF
clan_description: EGF superfamily
clan_comment: Members of this clan all belong to the EGF superfamily ...
Example query: Give me the literature references for clan ‘CL0001’
SELECT comment, order_added, pmid, title, author, journal
FROM clan, literature_reference, clan_lit_ref
WHERE clan.clan_acc = clan_lit_ref.clan_acc
AND clan_lit_ref.auto_lit = literature_reference.auto_lit
AND clan.clan_acc = 'CL0001'
Example output:
comment: NULL
order_added: 1
pmid: 3282918
title: Structure and function of epidermal growth factor-like regions in proteins.
author: Appella E, Weber IT, Blasi F;
journal: FEBS Lett 1988;231:1-4.
comment: NULL
order_added: 2
pmid: 11852228
title: Domain structure and organisation in extracellular matrix proteins.
author: Hohenester E, Engel J;
journal: Matrix Biol 2002;21:115-128.
Example query: Give me the database links for clan ‘CL0001’
SELECT db_id, comment, db_link, other_params
FROM clan_database_links, clan
WHERE clan_database_links.clan_acc = clan.clan_acc
AND clan.clan_acc = 'CL0001'
Example output:
db_id comment db_link other_params
SCOP NULL 57196
CATH NULL 2.10.25.10
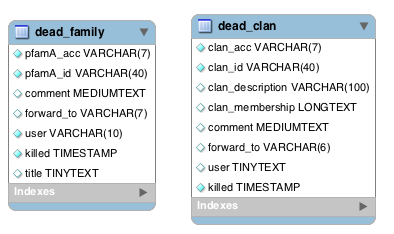
Dead families and clans¶
Sometimes we find that two or more Pfam-A families can be merged into a single family, which leads to the deletion of Pfam-A families. Likewise we might merge two clans together, which results in the deletion of a clan. The dead_family and dead_clan tables contain information about Pfam-A families and clans that have been deleted. These tables may be of use if you need to track what happened to the members of a particular family/clan that is no longer in Pfam.
Example query: Give me all of the information about ‘dead’ Pfam-A family ‘PF09410’
SELECT *
FROM dead_family
WHERE pfamA_acc = 'PF09410'
Example output:
pfamA_acc: PF09410
pfamA_id: DUF2006
comment: Merged into PF07143
forward_to: PF07143
user: jm14
killed: 2009-08-25 10:33:41
title: NULL
Example query: Give me all of the information about ‘dead’ clan ‘CL0152’
SELECT *
FROM dead_clan
WHERE clan_acc = 'CL0152'
Example output:
clan_acc: CL0152
clan_id: XI_TIM
clan_description: Xylose isomerase-like TIM barrel superfamily
clan_membership:
comment: Merged clan in to TIM_barrel clan
forward_to: CL0036
user: rdf
killed: 2009-06-22 17:47:17
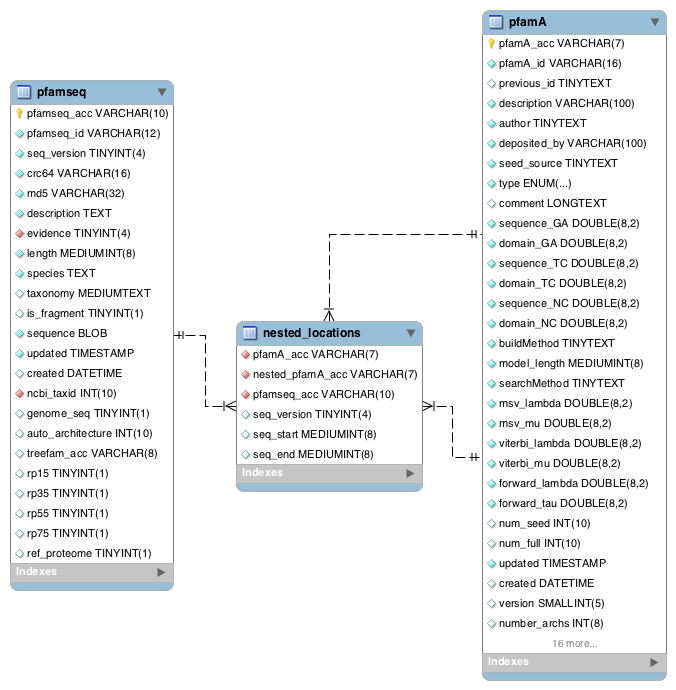
Nested domains¶
Some Pfam-A domains are disrupted by the insertion of another domain (or domains) within them. The domain that is inserted into another is known as a nested domain. The nested_locations table stores all the nested Pfam-A domains. It also stores the coordinates of the nested domain with respect to a sequence that is present in the seed alignment of the domain in which it nests.
Example query: Give me all of the nested domains and the domains in which they are nested
SELECT A.pfamA_id, B.pfamA_id AS nested_domain
FROM pfamA AS A, pfamA AS B, nested_domains
WHERE A.pfamA_acc = nested_domains.pfamA_acc
AND B.pfamA_acc = nested_domains.nests_pfamA_acc
Example output:
pfamA_id nested_domain
CusB_dom_1 HlyD_D4
CusB_dom_1 HlyD_D23
RTC RTC_insert
RtcB HNH_3
RtcB LAGLIDADG_3
RtcB Intein_splicing
RtcB Intein_splicing
Pro_dh EF-hand_7
LGT PDZ
HcyBio Fer4
...
Example query: Give me the nested data for the family IMPDH
SELECT pfamA_id, nested_pfamA_acc, pfamseq_acc, seq_version, seq_start, seq_end
FROM pfamA, nested_locations
WHERE pfamA.pfamA_acc = nested_locations.pfamA_acc
AND pfamA_id ="IMPDH"
Example output:
pfamA_id nested_pfamA_acc pfamseq_acc seq_version seq_start seq_end
IMPDH PF00571 Q21KQ8 1 155 271
Proteomes¶
As of Pfam 29.0, all sequences in the pfamseq table belong to a reference proteome, and therefore a complete proteome. Prior to Pfam 29.0 this was not the case. The complete_proteomes table contains statistics about the number of families and coverage. The tables in this section allow you to retrieve domain information about a particular species, or to retrieve all of the species which contain a particular Pfam domain.
Example query: Give me the Pfam summary for the human (Homo sapiens, ncbi taxid 9606) proteome
SELECT *
FROM complete_proteomes
WHERE ncbi_taxid=9606
Example output:
ncbi_taxid: 9606
species: Homo sapiens (Human)
grouping: Eukaryota
num_distinct_regions: 0
num_total_regions: 115682
num_proteins: 81828
sequence_coverage: 72
residue_coverage: 45
total_genome_proteins: 81828
total_aa_length: 29701382
total_aa_covered: 13467634
total_seqs_covered: 58973
Example query: Give me all the Pfam-A domains for the species ‘Arabidopsis thaliana’
SELECT r.pfamA_acc, pfamA_id, description, sum(number_domains)
FROM pfamA p, proteome_regions r
WHERE p.pfamA_acc=r.pfamA_acc
AND ncbi_taxid=3702
GROUP BY r.pfamA_acc
Example output:
pfamA_acc pfamA_id description sum(number_domains)
PF00004 AAA ATPase family associated with various cellular activities (AAA) 178
PF00005 ABC_tran ABC transporter 284
PF00006 ATP-synt_ab ATP synthase alpha/beta family, nucleotide-binding domain 13
PF00009 GTP_EFTU Elongation factor Tu GTP binding domain 48
PF00010 HLH Helix-loop-helix DNA-binding domain 233
PF00012 HSP70 Hsp70 protein 29
PF00013 KH_1 KH domain 106
...
Note: The ncbi_code for the species ‘Arabidopsis thaliana’ is 3702. This information can be found in the ncbi_taxonomy table.
Example query: Give me all of the protein sequences for the species ‘Arabidopsis thaliana’
SELECT pfamseq_acc
FROM pfamseq
WHERE ncbi_taxid = '3702'
Example output:
pfamseq_acc
A0A068FL09
A0A0A7EPL0
A0A140JWM8
A0A178U7J2
A0A178U7Y7
A0A178U807
A0A178U889
...
Example query: Give me all of the protein sequences from the species ‘Arabidopsis thaliana’ that belong to Pfam-A domain ‘PF00106’
SELECT pfamseq.pfamseq_acc
FROM pfamseq, pfamA_reg_full_significant
WHERE ncbi_taxid = '3702'
AND pfamseq.pfamseq_acc = pfamA_reg_full_significant.pfamseq_acc
AND pfamA_acc = 'PF00106'
Example output:
pfamseq_acc
A0A1I9LMA8
A0A1I9LNL3
A0A1I9LNL4
A0A1I9LNT2
A0A1I9LNT2
A0A1I9LQ55
A0A1I9LQ55
...
Data Files - Alignments, trees and HMMs¶
The seed, full, UniProtKB, NCBI, representative proteome and metaseq alignments are all stored as gzipped files in the database, as is the HMM for each family. Note that the NCBI and metaseq alignments may contain overlapping matches to Pfam-A families that belong to the same clan, however, the UniprotKB alignments (seed, full, uniprot and representative proteome sets) will not. This is because we have performed a clan filtering step on the UniProtKB data such that where there are overlapping Pfam-A matches within a clan, only the lowest E-value scoring match is included in the full alignment.