Summary¶
Proteins are generally comprised of one or more functional regions, commonly termed domains. The presence of different domains in varying combinations in different proteins gives rise to the diverse repertoire of proteins found in nature. Identifying the domains present in a protein can provide insights into the function of that protein.
The Pfam database is a large collection of protein domain families. Each family is represented by multiple sequence alignments and a profile hidden Markov model (HMM).
Each Pfam family, usually referred to as a Pfam-A entry, consists of a curated seed alignment containing a small set of representative members of the family, profile HMMs built from the seed alignment, and an automatically generated full alignment, which contains all detectable protein sequences belonging to the family, as defined by profile HMM searches of primary sequence databases.
Pfam entries are classified in one of six types:
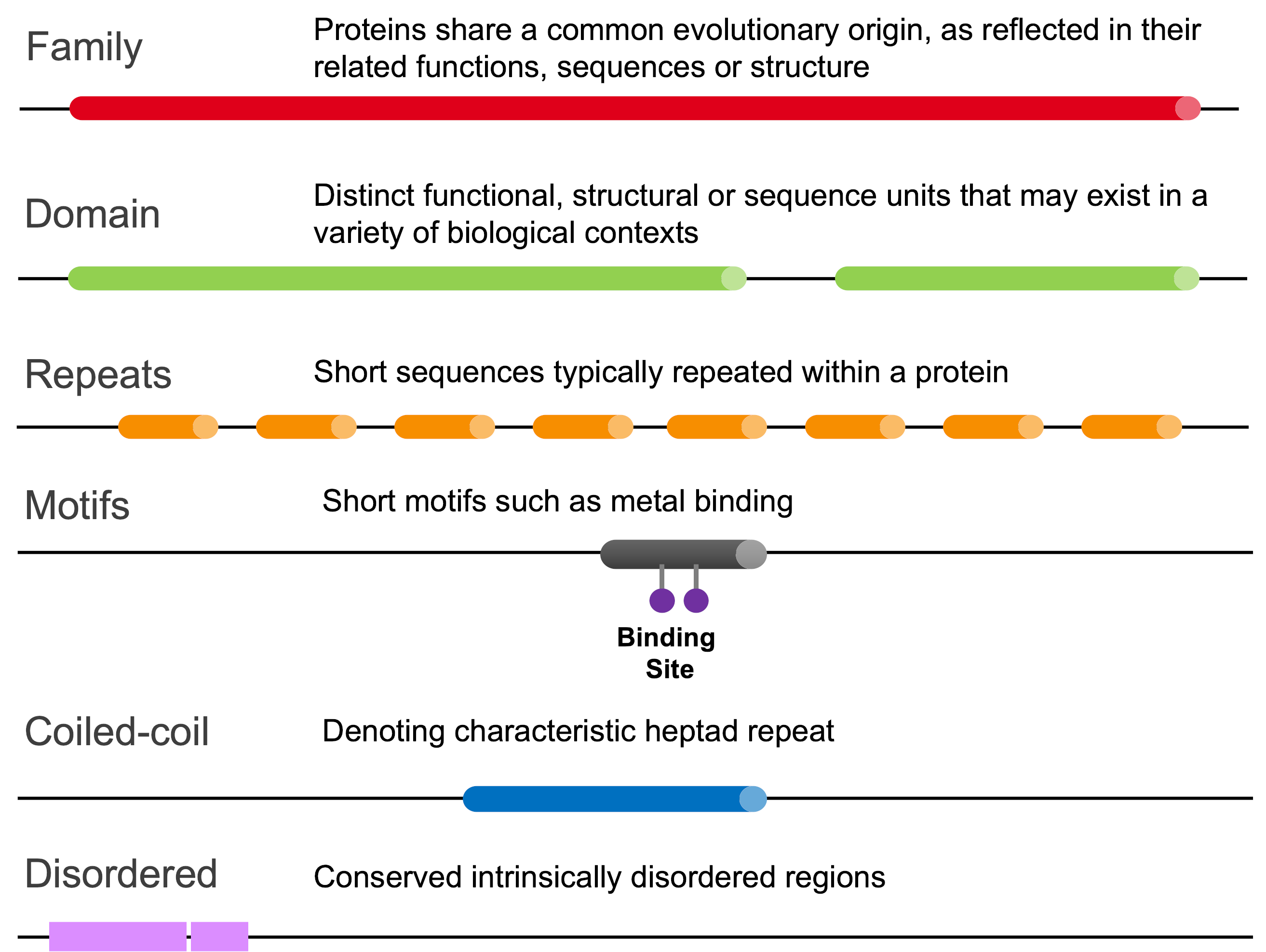
Types of Pfam entries.¶
Pfam clans¶
Structural properties are often more conserved than the underlying sequence. Therefore, a single profile HMM is often insufficient to model an entire, diverse, structural superfamily and related Pfam entries are sometimes grouped together into clans; the relationship may be defined by:
sequence similarity (whilst still originating from a common ancestor)
similarity of known three-dimensional structures
functional similarity
and/or similarity between their profile HMMs (as determined by algorithms such as HHsearch) similarity of sequence, structure or profile HMM.
The majority of Pfam Clans are groupings of domains and families.