Clan page organisation¶
If a Pfam entry is included in a Pfam clan this information will be displayed in the Overview tab in the Pfam entry page, next to Clan, below the Pfam short name, with a link to the corresponding clan page. More information about how clans are defined can be found in Summary.
Additionally, it is possible to browse through the Pfam clans by selecting Browse + By Clan/Set in the InterPro website menu and select Pfam in the database section.
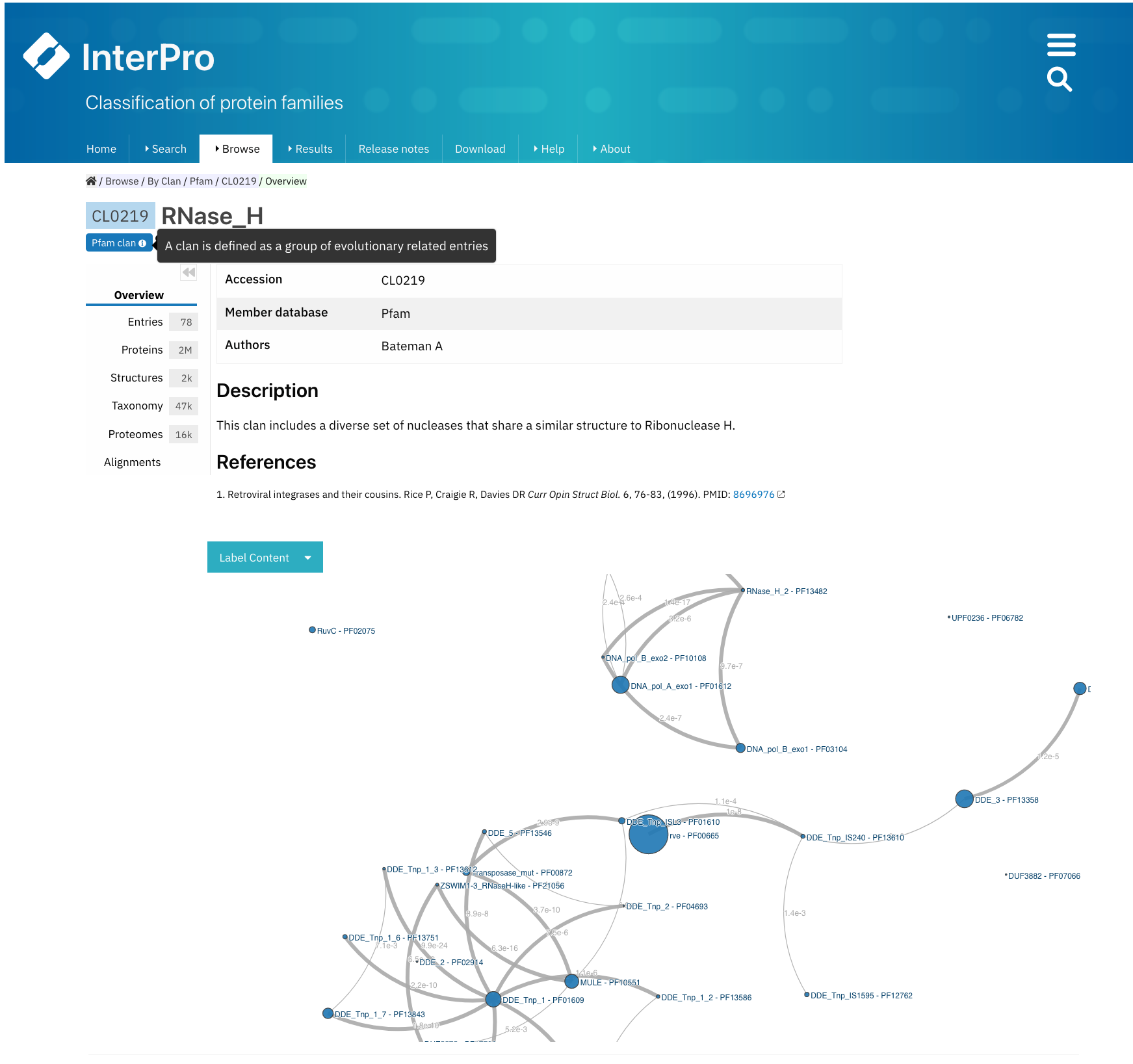
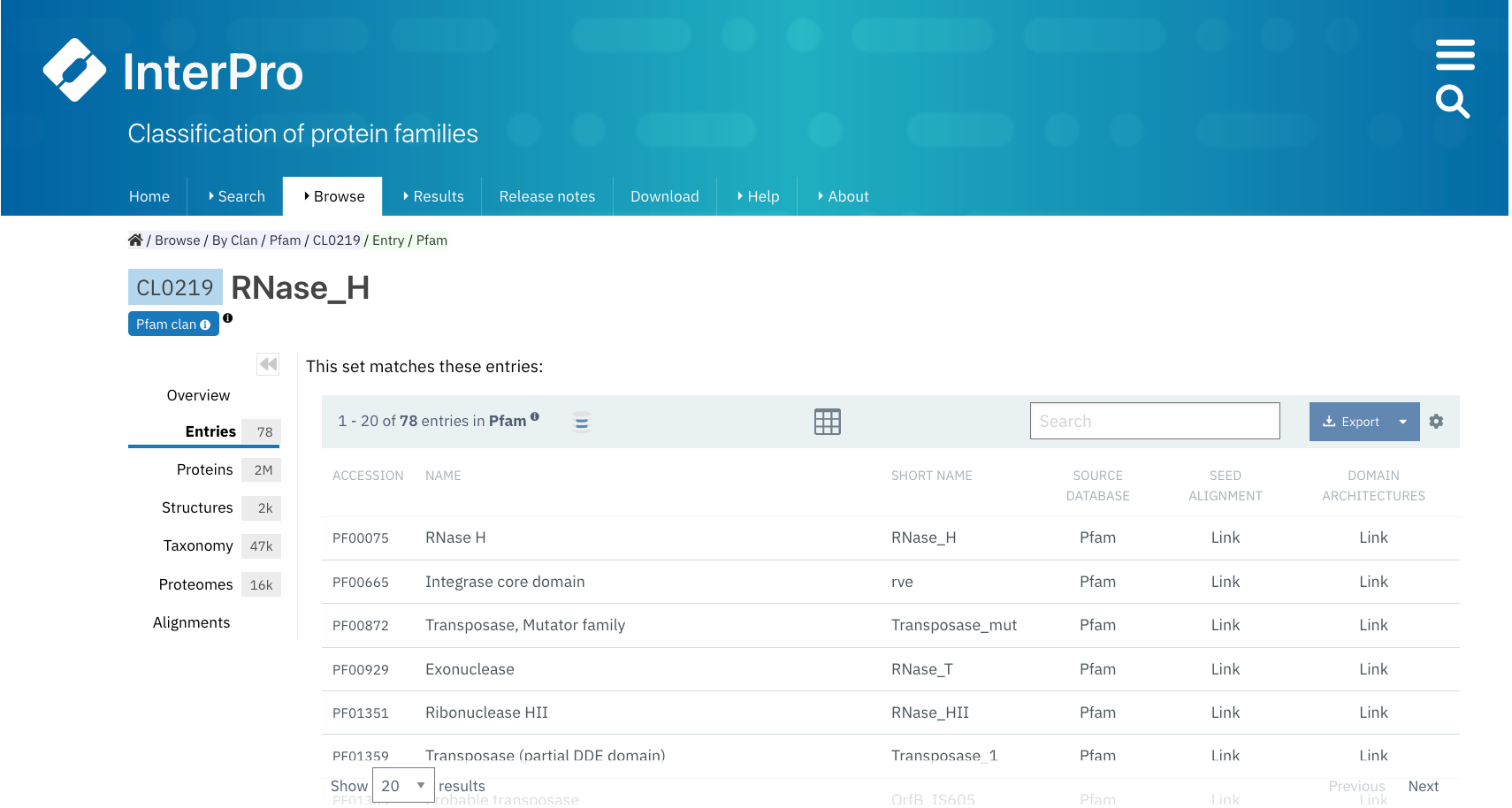
Example of a Pfam clan page (CL0219). All the tabs described below can be found on the left-hand side menu. The Overview tab is displayed by default.¶
In each Pfam clan page, different tabs with relevant information are available, the information they contain is described below.
Overview¶
The clan Overview tab is the default display, where the clan accession number, its short name and the author(s) are shown at the top. A description of the clan is displayed below, with the relevant literature references.
An interactive view of the Pfam entries included in the clan is also displayed, different label types can be chosen through the Label Content menu: Accession, Name and Short name.
Entries¶
The list of Pfam entries included in the clan is provided in this tab. For each entry, accession, name, short name and links to the entries SEED alignment and domain architectures pages are available.
Users can export this data in different formats, by clicking on the Export button, and customise the page settings, by clicking on the wheel icon.
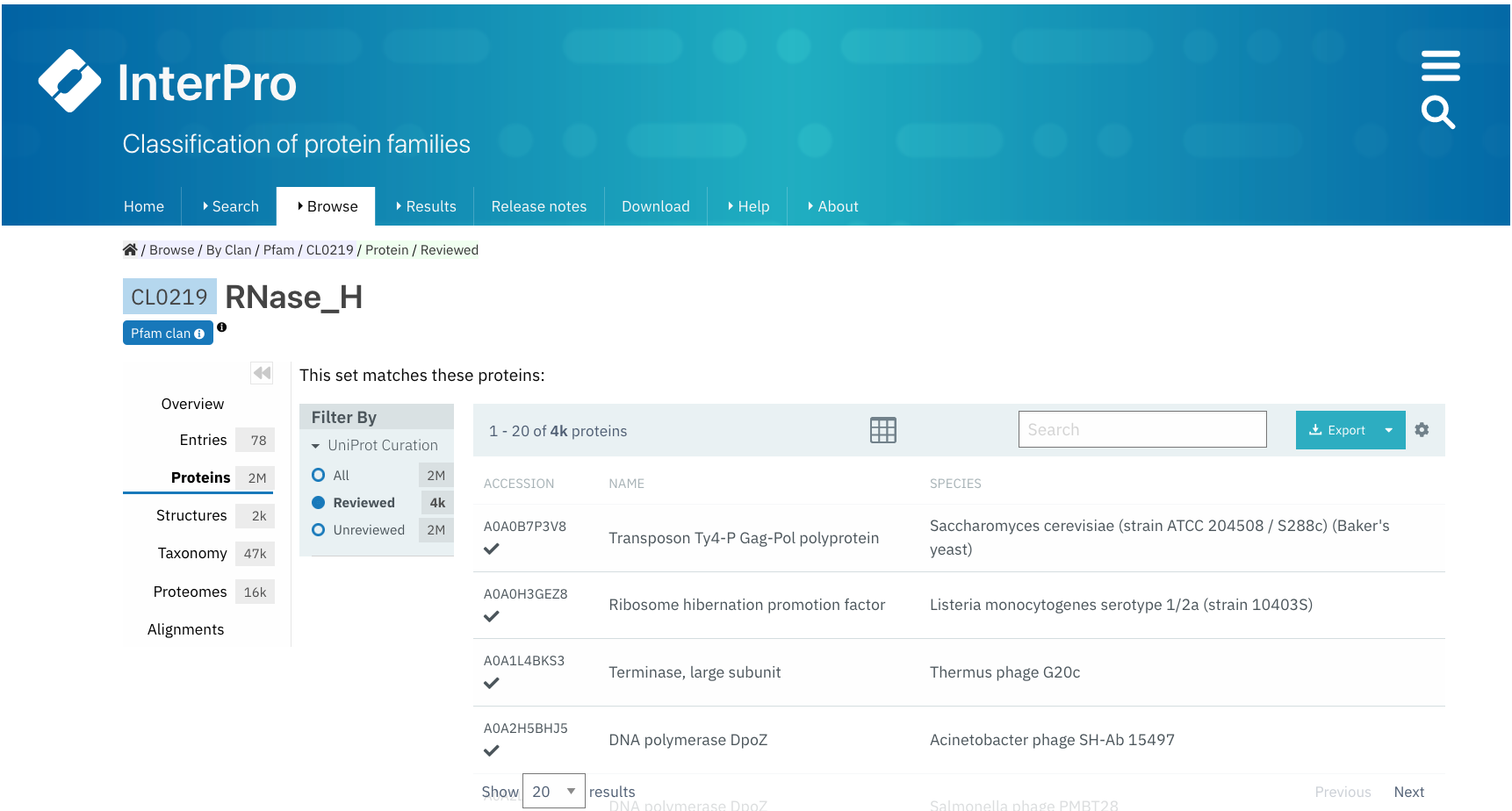
Proteins¶
The list of proteins matching any Pfam entry belonging to the clan is displayed in this tab. The view can be customised to show:
All proteins (from the whole UniProtKB database).
Only Reviewed proteins (from SwissProt - manually curated).
Only Unreviewed proteins (from TrEMBL - derived from public databases automatically integrated into UniProt).
For each protein, the corresponding protein page in InterPro can be accessed by clicking on the protein accession or name, and the InterPro taxonomy page can be accessed by clicking on the species name.
Users can export this data in different formats, by clicking on the Export button, and customise the page settings, by clicking on the wheel icon.
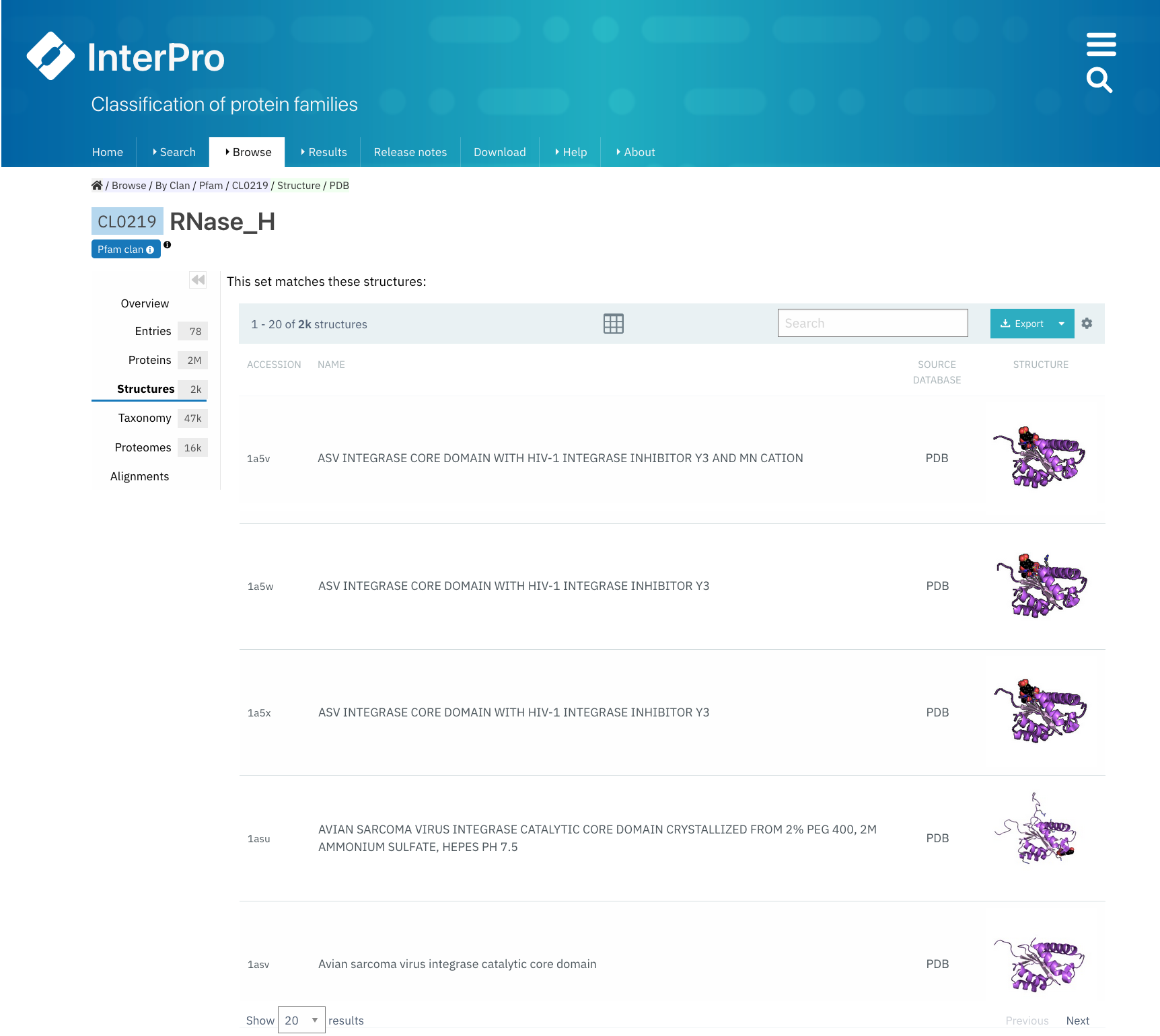
Structures¶
This tab displays a list of all the PDB structures linked to the proteins matching any Pfam entry belonging to the clan. For each structure, you can see the PDB accession and the name of the structure in PDB.
By clicking on a PDB accession, name or small image of the structure, a view of the corresponding InterPro structure page that summarises all of the entries of Pfam and other databases and resources for each chain of the structure will be displayed in a protein sequence viewer.
The position of each entry within the overall 3D structure can be visualised by choosing the Pfam entry of interest in the drop-down list Highlight Entry in the 3D structure or by clicking on the bar corresponding to the entry match in the protein sequence viewer. Additionally, links to similar PDB viewers and cross-references to other structural databases are provided in the External links section.
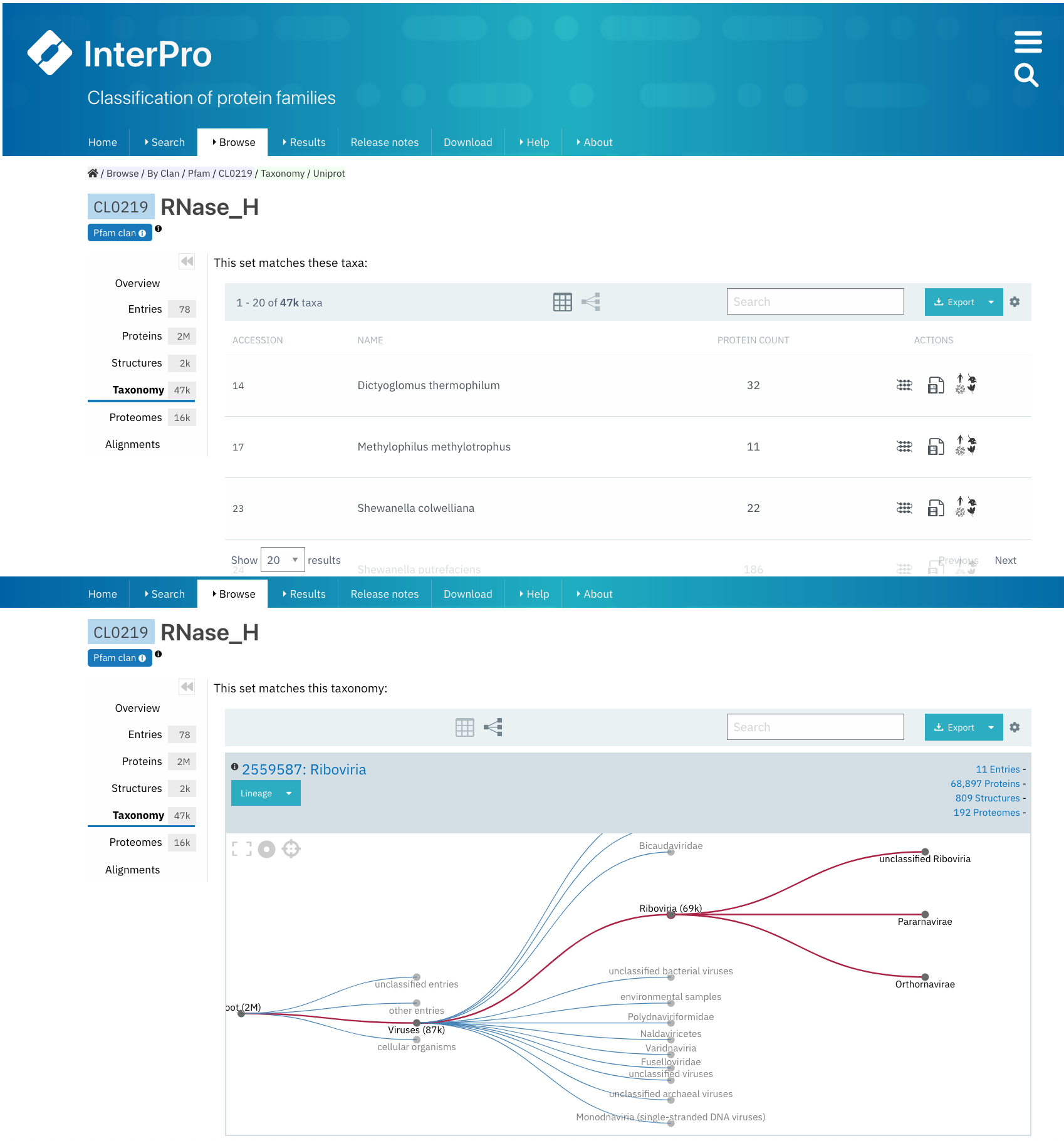
Taxonomy¶
This tab shows by default a list of all the species that the proteins matched by any Pfam entry of the clan belong to.
These data can also be seen as a tree. These visualisation options can be chosen from the icon panel above the list. All this information can be downloaded in different formats.
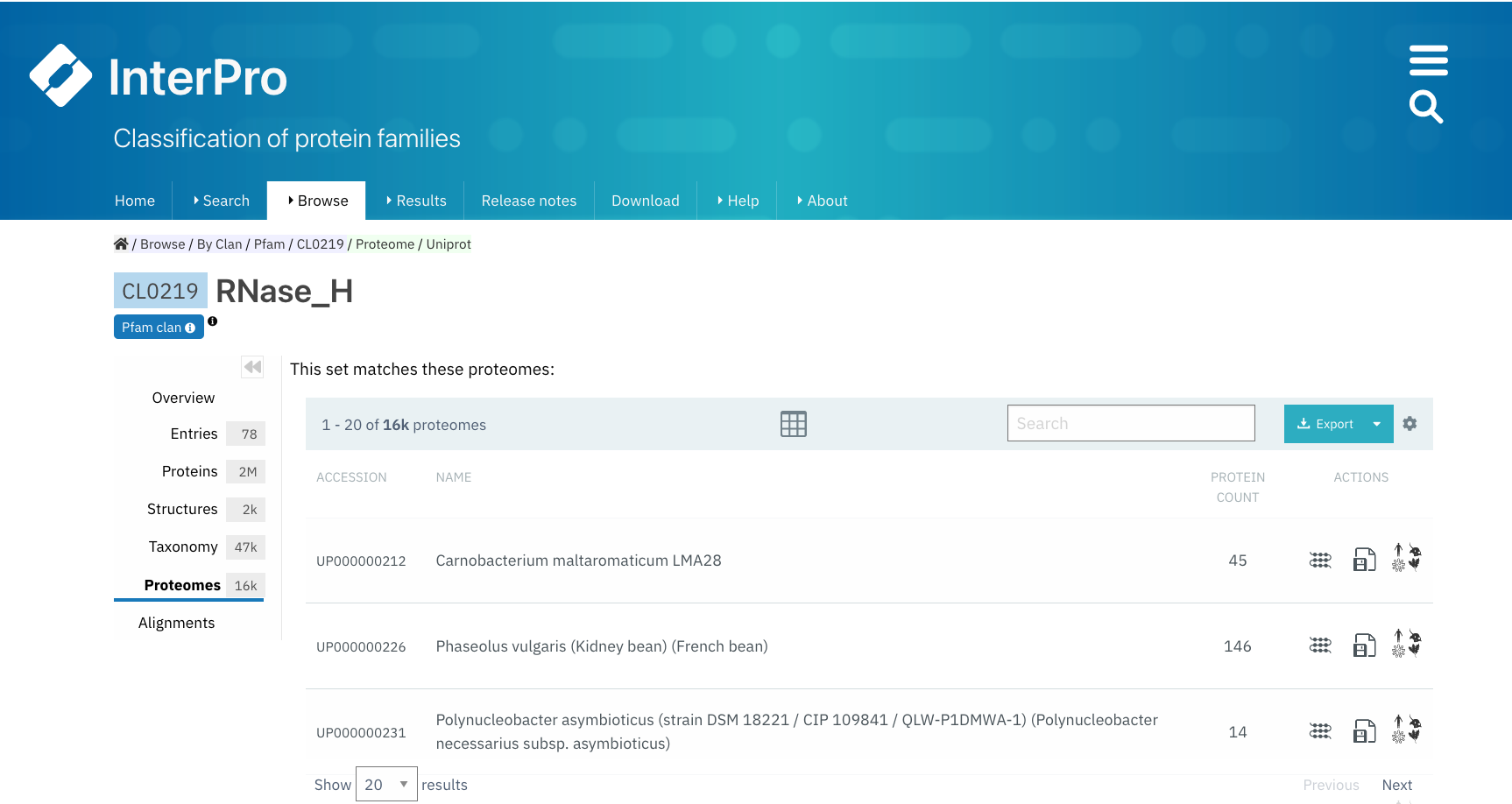
Proteomes¶
A list of the reference proteomes matched by any Pfam entry belonging to the clan is displayed in this tab. For each item in this list the Proteome ID (which is a link to the Proteome page in InterPro), the name of the species carrying this proteome and the number of proteins in this proteome that match the entry are displayed. From the Actions column, users can also access a list of these proteins by clicking the first icon (View matching proteins), download the data in different formats or View proteome information.